Future Ready
Northwestern’s new Center for Pathogen Genomics and Microbial Evolution is laying the groundwork to prevent future pandemics.
by Bridget Kuehn

Simons, Egon Ozer, MD, PhD, and Judd Hultquist,
PhD — the team behind the new Center for
Pathogen Genomics and Microbial Evolution
Just as the COVID-19 pandemic started to upend many Chicagoans’ lives in March 2020, a team of Northwestern scientists made a startling discovery. By sequencing the genome of SARS-CoV-2, the virus that causes COVID-19, in samples collected from patients in Chicago, they discovered three distinct strains were already circulating in the city.
“The virus had already spread at least three different ways across the globe to arrive in Chicago,” says Egon Ozer, MD, PhD, ’08, ’12 GME, assistant professor of Medicine in the Division of Infectious Diseases. The team also discovered that some of the genetic changes in the viral genome increased the amount of virus in the airways of infected people, making them more likely to spread the virus to others and hastening the spread of the virus in the area.
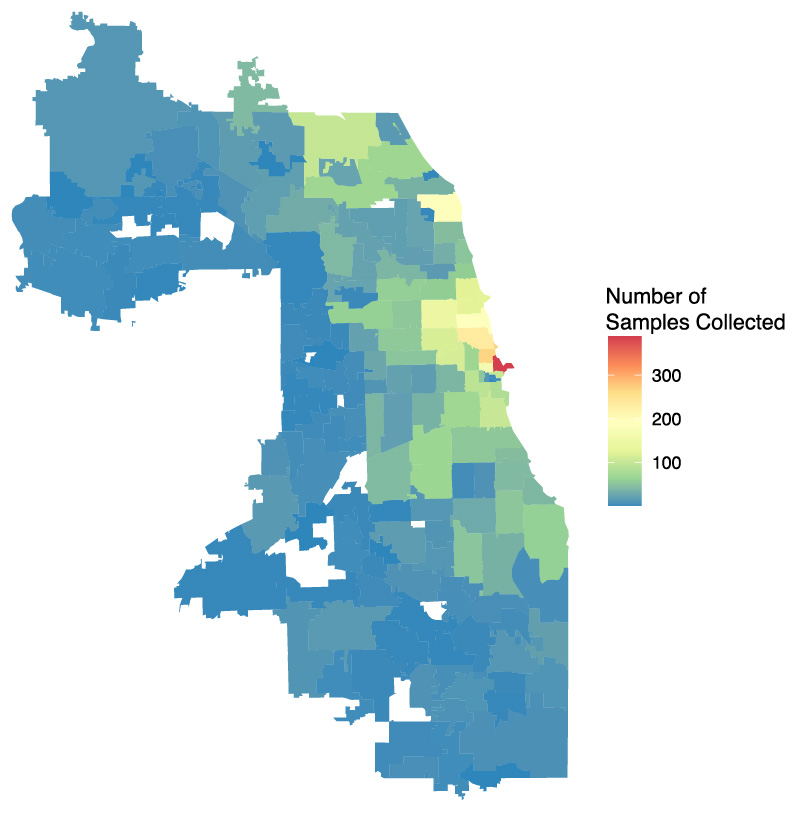
from Cook County,
Illinois
The CPGME has collected viral specimens from individuals with COVID-19 across Chicago and Cook County since March. Genomic sequencing of some of these specimens helps the team understand how the SARS-CoV-2 virus is spreading and changing in the area.
These discoveries were made possible by a nimble collaboration that included Ozer, an expert in bacterial genome sequencing, virologist Judd Hultquist, PhD, assistant professor of Medicine in the Division of Infectious Diseases, bioinformatics expert Ramon Lorenzo-Redondo, PhD, a research assistant professor of Medicine in the same division, and laboratory coordinator and biomedical researcher, Lacy Simons. During the course of the pandemic, this team has overseen the sequencing of more than 1,800 samples of SARS-CoV-2 from patients in the Chicago area.
To continue helping with SARS-CoV-2 global surveillance efforts — and begin monitoring for new potential viral and microbial threats — the medical school has launched a new Center for Pathogen Genomics and Microbial Evolution (CPGME) in the Institute for Global Health, with Ozer serving as director.
“When COVID-19 came along, it really crystallized the power of whole-genome sequencing of pathogens,” says Ozer. “It’s imperative for understanding where a pandemic comes from, how it is spreading, how it can be stopped, and how to prevent the next one.”
Rapid Response Team
The new center will provide the expertise and analytic tools required to facilitate ongoing analysis of the genetic changes occurring in SARS-CoV-2, as well as help to identify potential new pandemic threats. It will also use genomics to better understand what drives the emergence of antimicrobial resistance and how to stop it.
“Pathogens are always changing,” explains Hultquist, associate director of the new center. “Viruses change very quickly, bacteria can change very quickly, and by continuing to track the changes that they’re accumulating in their genomes we can map how they’re being transmitted within a community or even within a workplace. But until now, Northwestern hasn’t had a dedicated center with a focused investment in these microbial sequencing techniques and technologies.”
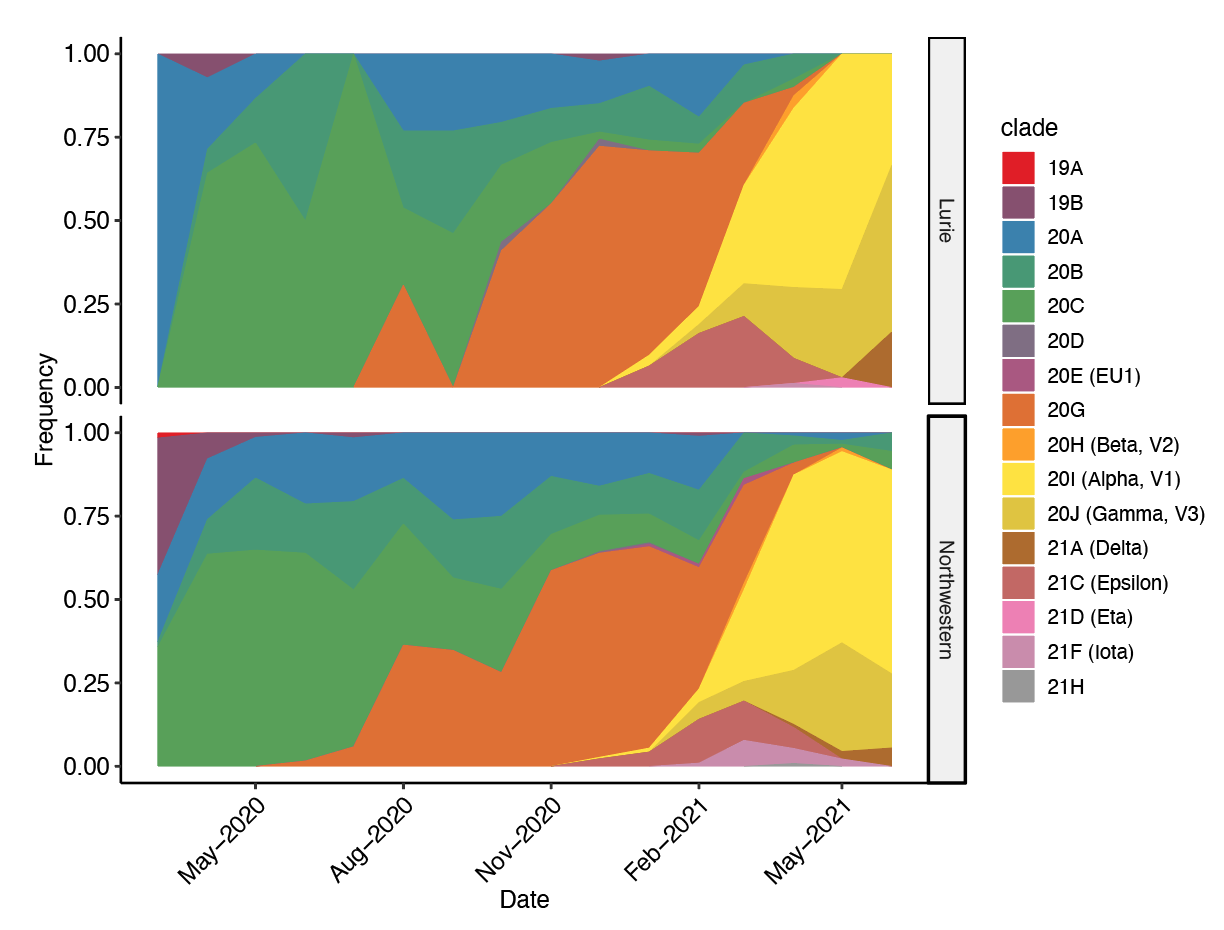
This chart shows monthly proportions of the most common SARS-CoV-2 clades found among the sequences obtained by CPGME. The distribution of clades within adult and pediatric populations are very similar, indicating that there are not major differences between the two populations. By tracking the viral variants, CPGME can study the population dynamics occurring in the Chicago area and compare it to the global epidemic. This information is key to designing public health strategies.
In addition to providing an in-house resource, the center will also continue to foster the collaborations between basic scientists, clinicians, and the public health departments that have been essential during this pandemic.
“When entirely developed, this approach can be one of the most powerful resources to inform public health measures against infectious pathogens and ultimately prevent future epidemics,” says Lorenzo-Redondo, who is CPGME’s bioinformatics director. “We want this center to build expertise in pathogen genomics and evolution to generate a network of investigators who are capable of managing current and future infectious disease threats.”
That network will extend across the globe through partnerships with institutions in Nigeria and Pakistan, where few pathogens have undergone genomic sequencing. Ozer explains that as sequencing becomes less expensive and more user-friendly, access to the technology is expanding. However, many organizations don’t yet have the bioinformatics expertise necessary to analyze microbial genomics data, which requires different techniques than human or animal genomics.
“What we think we can contribute by partnering with scientists in Pakistan and Nigeria is to build up their capacity to perform these analyses, help them identify emerging pathogens, and help them understand transmission in their communities,” Ozer says.
Ongoing Surveillance
Those international partnerships and the CPGME team’s ongoing surveillance of changes in the SARS-CoV-2 genome are essential to helping identify new variants that may pose enhanced risks. For example, collaborations have already led to the characterization of a concerning variant circulating in West Africa, according to Hultquist.
“Until we really have a handle on what kind of variation there is in the virus worldwide, we are not going to get the pandemic under control,” he says.
We want this center to build expertise in pathogen genomics and evolution to generate a network of investigators capable of managing current and future infectious disease threats.
Ramon Lorenzo-Redondo, PhD
The team is also continuing to track changes in the virus locally. Last summer, they identified a new strain that went on to drive the winter peak in infections. They were also the first group to identify the arrival of the Alpha variant, a more transmissible version of the virus that first emerged in the United Kingdom and documented how it quickly became the dominant strain in Chicago.
“Chicago is a very interesting place to study the virus because it has one of the most connected airports in the world,” Lorenzo-Redondo says.
As other versions of the virus, such as the Delta variant, emerge in the U.S. and around the world, the team will help to monitor their spread in Chicago.
“As some variants have the potential to transmit infection more easily, cause more severe disease, or be less susceptible to vaccines, it is important to identify and track these infections to help guide clinical and public health responses,” says Ozer.
The center is also working to help answer a pressing question on the mind of many Chicago parents as schools prepare to resume in-person instruction in the fall: Is it safe? That project is being led by Larry Kociolek, MD, ’14 GME, assistant professor of Pediatrics in the Division of Infectious Diseases, who has been working with the CPGME team to sequence SARS-CoV-2 samples from children attending school to determine if spread is happening within schools or if children are instead being infected in the community.
As more people in the U.S. and around the world are vaccinated, the team is also looking for potential changes in the virus that might let it evade the immune protection offered by vaccination, Ozer says. This is particularly important because the slow rollout of vaccines around the world and the uneven vaccine coverage in the U.S. has created pockets with high rates of infection — ideal circumstances for the virus to develop new variants.
“It is important for us to continue the SARS-CoV-2 genomic surveillance,” Ozer says. “We are hopefully rounding the corner in the pandemic, but it is certainly not over yet.”
Tackling Future Threats
As the CPGME team continues to monitor the ongoing changes in the SARS-CoV-2 virus, they are already broadening their sights to tackle one of the biggest ongoing challenges in medicine: antimicrobial resistance. Disease-causing bacteria and fungi that have become immune to many currently available medications cause about 2.8 million hard-to-treat infections and more than 35,000 deaths in the U.S. each year, according to the U.S. Centers for Disease Control and Prevention. This is also a growing global threat, claiming more than 700,000 lives each year — a number that could grow to 10 million people a year by 2050, according to a review commissioned by the United Kingdom.
It is important for us to continue the SARS-CoV-2 genomic surveillance. We are hopefully rounding the corner in the pandemic, but it is certainly not over yet.
Egon Ozer, MD, PhD
To help blunt this threat, the team will partner with clinicians and the clinical microbiology laboratory at Northwestern to screen for drug-resistant microbes in its hospitals and the surrounding community, Ozer says. The hope is to help identify how patients are infected and to develop prevention strategies. For example, in the last decade a fungus called Candida auris, which is often resistant to multiple antifungal drugs, has emerged. It can cause life-threatening infections in hospitalized or severely ill patients and is very difficult to treat. The team is using genomics to track these fungi to better understand how they spread in hospitals, how they develop resistance to antimicrobial drugs, and if they are able to share their drug-evading strategies with other organisms.
“We are hoping to better understand the mechanisms of antimicrobial resistance,” Ozer says.
Mehreen Arshad, MBBS, assistant professor of Pediatrics in the Division of Infectious Diseases, is also leading a pilot study funded by the Northwestern Buffett Institute for Global Affairs to develop a way for the CPGME and its partners in Pakistan to share data and specimens of drug-resistant microbes. The goal is to better understand how both local and global factors influence the use of antimicrobial drugs by clinicians and how these behaviors may reduce or contribute to the emergence of drug-resistant organisms.
“Sequencing is one of the very few tools that can powerfully attack the problem of antimicrobial resistance so that we can understand it better and come up with new solutions,” Hultquist says. “Transitioning from our work in viruses to our work on other microbes is going to be really important going forward.”
A CLOSE EYE ON INFECTIOUS DISEASE
From antimicrobial resistance to the quest for a cure to HIV/AIDS, these Feinberg scientists are taking an expansive view of these microscopic pathogens.
Steven Wolinsky, MD, ’82 GME, the Samuel Jefferson Sackett Professor of Infectious Diseases, and Karla Satchell, PhD, professor of Microbiology-Immunology and co-director of the Center for Structural Genomics of Infectious Diseases, have spent years studying dangerous pathogens.
Wolinsky’s research focuses on the evolutionary mechanisms underlying host-pathogen interactions. Understanding the evolutionary arms race between viruses and the immune system is the best way to identify therapeutic pathways, Wolinsky says.


“Human and virus populations are constantly under selective pressures to maintain a competitive advantage over one another,” Wolinsky says. “Mutations that alter the genes in host networks involved in viral replication and early immune defense may underlie the lack of viral control in some people and the apparent resistance to infection in others.”
Satchell, on the other hand, focuses on bacteria. Bacteria cause enormous morbidity and mortality every year, and the rise of antimicrobial-resistant strains threaten to upend hospital care and unleash a new wave of pathogens on the planet.
Satchell investigates secreted protein toxins, which are released during infection to damage host cells. Satchell’s research delves into their contribution to infection in cholera, seafood-associated septicemia, and necrotizing fasciitis. Recently, Satchell investigated the structure of SARS-CoV-2, searching for weak spots in the virus’ armor.
“Microbes manipulate our cells in many fascinating ways to block our defense systems so the infection can take hold,” says Satchell, who is also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University. “Our work on how toxins of bacteria accomplish infection helped us understand how SARS-CoV-2 also circumvents our defense strategies, as the experimental techniques and concepts are similar.”