Decoding the Epigenome
Investigators in Feinberg’s Department of Biochemistry and Molecular Genetics work to understand influences on gene expression — with new drug treatments on the horizon
By Emily Ayshford
When the first complete human genome was sequenced in 2003, many thought understanding DNA would herald a new paradigm in medicine. More than 20 years later, scientists in Feinberg’s Department of Biochemistry and Molecular Genetics and Simpson Querrey Institute for Epigenetics are at the forefront of understanding epigenetics — how environmental influences impact the cellular processes that regulate gene expression — and how those processes can lead to disease.
“We are in the business of discovering critical factors in the pathways that control gene expression programs in cancer and other diseases,” said Ali Shilatifard, PhD, the Robert Francis Furchgott Professor and chair of the department. “Once we have identified these factors, we want to develop drugs to target them and move them into the clinic to save lives. That is our ultimate goal.”
At the heart of epigenetics is chromatin, the complex of DNA and the proteins that control its storage and access. Within chromatin are histones, regulatory proteins that play a key role in gene expression. Chemical alterations to histone proteins can facilitate gene expression by allowing transcription factors and enzymes to interact with DNA, while other histone alterations can prohibit these interactions to limit gene expression. Access to DNA is also regulated by DNA methylation, a process in which methyl groups are added to DNA without affecting the DNA sequence itself. Adding to this complexity, gene expression is also epigenetically regulated by non-coding RNAs.
In recent years, the field of epigenetics has been boosted by new technology such as gene editing tool CRISPR/Cas9, and scientists have been emboldened by research that has shown the key to treating diseases like cancer lies not in manipulating genes, but in better understanding gene expression.
We are in the business of discovering critical factors in the pathways that control gene expression programs in cancer and other diseases. Once we have identified these factors, we want to develop drugs to target them and move them into the clinic to save lives. That is our ultimate goal.
Ali Shilatifard, PhD
FINDING NEW TREATMENTS FOR CANCERS
Shilatifard’s team has found that mutations causing defects in a family of transcriptional and epigenetic regulators called COMPASS are linked to a variety of cancers, including bladder cancer.
“For a long time, investigators primarily focused on genes thought to drive cancer,” said Shannon Lauberth, PhD, associate professor of Biochemistry and Molecular Genetics. “However, many of these genes have proven challenging to target therapeutically. What has become increasingly evident is the dynamic and reversible nature of the epigenome, which allows for the reprogramming of gene expression. This offers a unique opportunity to inhibit tumor-promoting gene expression programs. By targeting epigenetic regulators, we can develop compounds with the potential to effectively disrupt these programs.”
In collaboration with Zibo Zhao, PhD, assistant professor of Biochemistry and Molecular Genetics, and physicians at Northwestern Medicine, Shilatifard discovered that an FDA approved drug called pemetrexed — originally developed for lung cancer — could be used to treat tumors with these COMPASS mutations. The results were published in the Proceedings of the National Academy of Sciences (PNAS), and a clinical trial to test pemetrexed is now being launched at Northwestern Medicine.
Other investigators have their eyes on similarly hard-to-treat cancers. Lung cancer is the leading cause of cancer deaths worldwide. Small-cell lung cancer, in particular, has a low survival rate, with no good treatment options.
Lu Wang, PhD, assistant professor of Biochemistry and Molecular Genetics, discovered that inhibiting a chromatin remodeling complex associated with the gene POU2AF2 could decrease tumor growth in small-cell lung cancer patients.
The gene that Wang discovered collaborates with the chromatin remodeling complex to recruit factors to maintain its genetic expression. Targeting these epigenetic factors that are recruited by POU2AF2 could be a potential treatment option. In fact, in a paper published in Nature Communications, the team showed that a commercially available drug that inhibits this chromatin remodeling complex significantly reduced tumor growth.

“The field of epigenetics is still pretty new, so 95 percent of commercial drugs used to treat cancer are not epigenetic drugs,” Wang said. “Now we know how important epigenetics is to cancer. At Northwestern, our research buildings are physically connected to the hospital. It’s a great platform for both basic science research and clinical translation to get new epigenetic drugs into the clinic.”
Another subtype of lung cancer, non-small cell lung cancer, accounts for about 85 percent of cases. About 20 percent of these cases have the LKB1 genetic mutation, but there are no effective targeted therapies for this cancer type.
Lillian Eichner, PhD, assistant professor of Biochemistry and Molecular Genetics, knew that the LKB1 protein, encoded by the STK1 gene, behaved differently, and that tumors inactivated for LKB1 have a distinct transcriptional signature. She and her team studied epigenetic factors and found that a certain histone deacetylases protein called HDAC3 was critical to the growth of LKB1-mutant tumors.
They tested the drugs entinostat (an HDAC inhibitor in clinical trials) and trametinib (an FDA-approved inhibitor of a different class of enzymes) in a mouse model. They knew that lung tumors become resistant to trametinib, but their work revealed that co-treatment with entinostat could help overcome that resistance. They found that mice given both drugs had 79 percent less tumor burden in their lungs than untreated mice after six weeks of treatment.
“We found that HDAC3 is really important for controlling how a tumor responds to treatment,” Eichner said of the work, which was published in Science Advances. “And it is a good target; it’s druggable.”
In a follow-up study published in PNAS, the team also identified a role for HDAC3 in controlling immune cells inside the tumor, which could prove to be an additional benefit provided by targeting HDAC3 for lung cancer treatment. “Our studies show that if we understand what a protein is doing, if we know that it supports tumor survival, we can reverse that with drug therapy,” she said.
UNDERSTANDING FACTORS AND REGULATORS TO FIND NEW DRUG TARGETS
Despite all the knowledge that scientists have about the genomes of cancer cells, many cancers remain difficult to treat. That’s because cancer cells can adapt to treatments, circulate within the body, and live in a tissue far from where they originated.
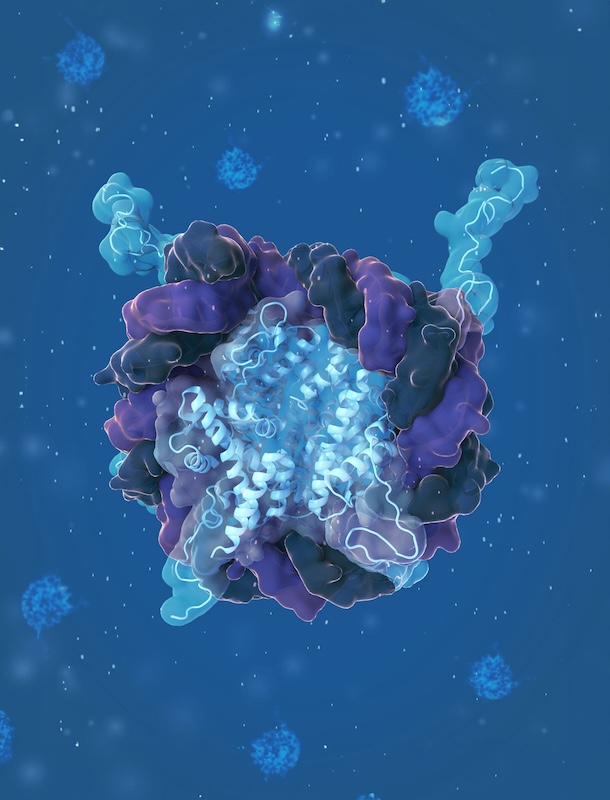
Marc Mendillo, PhD, associate professor of Biochemistry and Molecular Genetics, has worked to find the transcriptional factors that make this resilience possible. He and his team discovered that heat shock factor 1 (HSF1), a transcription factor known for inducing a cell’s heat shock response, also drives a gene expression program in cancer cells that promotes resilience and enables tumor progression.
Image courtesy of Brianna Monroe, MS
They then took that research further, discovering that HSF1 activity can be regulated by one of its transcriptional targets — a protein called the Jumonji domain-containing protein 6 (JMJD6). The work, published in PNAS, is another link that shows how cellular regulators enable the survival of cancers.
“We are looking for new drug targets that may not have been on people’s radars,” Mendillo said. “We want to lay out proof-of-principle studies that get people excited about these targets. We want to do something that impacts patients sooner rather than later.”
Identifying novel targets that others may have overlooked is also a key focus for Lauberth. “I’m passionate about the process of piecing together the puzzle of the epigenome to gain a deeper understanding of how it functions,” she said.
Lauberth’s lab is dedicated to studying the dynamic nature of the epigenome, with a particular focus on a class of important epigenetic regulators known as downstream-of-gene (DoG) RNAs. Unlike most RNAs, DoGs are not translated into proteins. Instead, they are produced when the transcription machinery continues past the normal end point of a gene, often in response to stressors such as viral infections or heat shock.
“Researchers have largely overlooked these molecules because they were thought to have no known function,” Lauberth said. “However, we’ve found that certain DoGs are produced in various types of cancer and are associated with reduced patient survival. Our goal is to understand if we can target these DoG RNAs to disrupt their expression and mitigate the severity of cancer disease and progression.” Her team’s findings were published in Science Advances.
Seeing this research through to its full potential and ultimately helping patients in a more impactful way — that’s the ultimate goal.
Shannon Lauberth, PhD
Although RNA-based therapeutics remain a relatively new approach to treating disease, Lauberth remains optimistic. Her team has already conducted a high-throughput screen to identify compounds that could target a key regulator of DoG RNA. “Seeing this research through to its full potential and ultimately helping patients in a more impactful way — that’s the ultimate goal,” she said.
While much of the department’s research centers on cancer, investigators are discovering that many of the epigenetic pathways that they study are also altered in a variety of other diseases. “That’s what makes this work so exciting,” Lauberth said.
ANOTHER PIECE OF THE PUZZLE: CELL METABOLISM
Within the Department of Biochemistry and Molecular Genetics, investigators are also conducting groundbreaking work on cell metabolism — which could ultimately influence epigenetic events.
Issam Ben-Sahra, PhD, associate professor of Biochemistry and Molecular Genetics, recently discovered how cellular metabolism — the chemical reactions that provide energy for a cell — fluctuates in response to changes in levels of metabolites called pyrimidines.
When levels of pyrimidine metabolites (which are used by cells to make DNA and RNA) go down, it reduces the cell’s ability to create energy. The results, published in Science, essentially rewrite what is known about how cells function.
Building on these findings, Ben-Sahra is now working with other investigators within the department to study how metabolites like this can fuel epigenetic changes. “We are working to link epigenetic mechanisms and metabolism,” he said. “If we disrupt the epigenetic landscape, how does cellular metabolism respond to that? This is a largely unexplored area, and we are eager to shed light on this black box.”
EXPLORE MORE
DEDICATED TO UNDERSTANDING THE NATURE OF THE EPIGENOME
In 1996, Shilatifard showed for the first time that transcriptional elongation — the process of extending the RNA strand synthesized from DNA — is a key step in regulating gene expression. If this step goes awry, he showed, it can cause leukemia.
After his groundbreaking work in understanding gene transcription, Shilatifard arrived at Feinberg in 2015 ready to build a department of investigators to study the molecular and genetic bases of human disease.
A ‘REVOLUTIONARY’ TECHNIQUE TO STUDY PROTEINS
Gene targeting and editing tool CRISPR/Cas9 has pushed the epigenetics field forward over the last decade. But Bercin Kutluk Cenik, MD, PhD, a former postdoctoral fellow in the lab of Ali Shilatifard who is now a research assistant professor, wanted to take that technology a step further. Over the past five years, she has worked to combine the gene-targeting feature of the CRISPR/Cas9 technology with proximity labeling, a technique to identify interactions between proteins and RNA.
The result is TurboCas, which allows investigators to identify and label which transcription factor proteins are involved in regulating the expression of a specific gene. TurboCas specifically labels proteins near the targeted DNA sequence. This helps researchers understand how proteins interact with each other and with chromatin at specific sites in the genome to control gene activity.
“The precision of it is what makes it revolutionary,” Cenik said. The team will now share the technique with the scientific community, with the ultimate goal of creating an interactive database to map how transcription factors interact across the genome.